Written by: Alyssa Sevilla
Clinically reviewed by: Robert Philibert
Causality refers to a relationship where a change in one factor directly leads to a change in another aspect. In contrast, correlation refers to a relationship where two factors are related or associated, but a change in one does not necessarily cause a difference in the other. In other words, causality implies a cause-and-effect relationship, while correlation simply indicates a relationship between two variables. In epigenetics, it is essential to distinguish between causation and correlation because many epigenetic changes may be associated with a disease or trait but not necessarily be the cause of it.
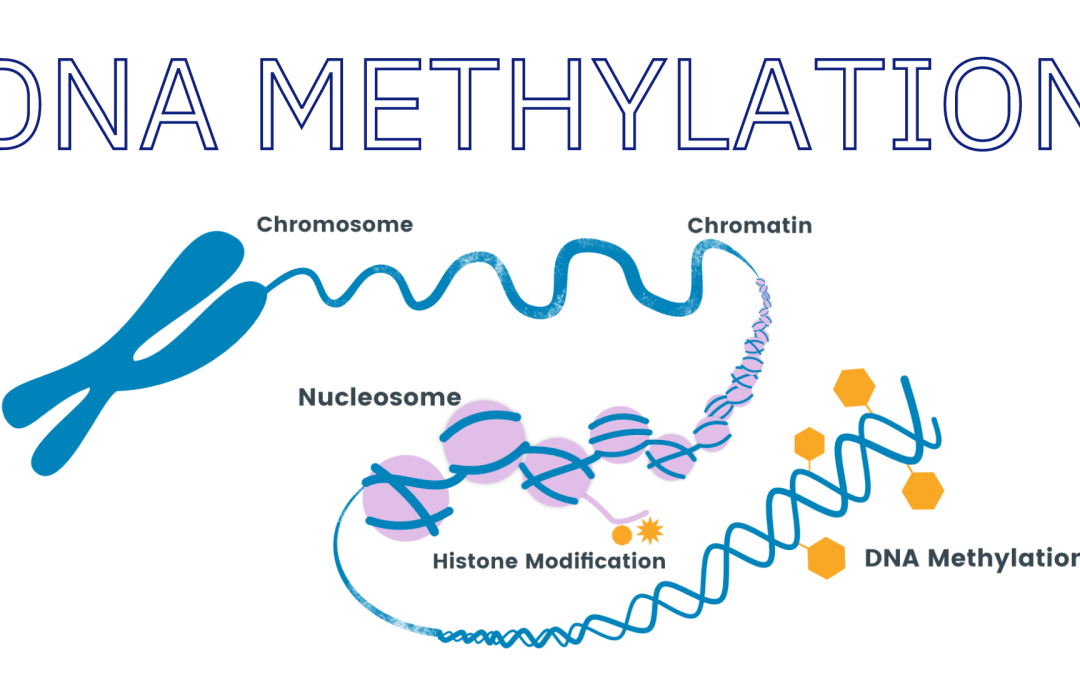
DNA methylation is an epigenetic modification that involves adding a methyl group to a cytosine base in DNA. Changes in DNA methylation patterns have been associated with various diseases, including cancer, neurological disorders, and cardiovascular diseases. By studying these changes, researchers can gain insight into the molecular mechanisms underlying these diseases and develop new diagnostic and therapeutic approaches.
For example, changes in DNA methylation patterns can be used as markers for the early detection of cancer. In some cancers, specific genes are hypermethylated, meaning that they have a higher level of methylation compared to normal cells. This can lead to the repression of gene expression, which can contribute to the development of cancer. By detecting these changes in DNA methylation, researchers can identify individuals at risk for cancer and monitor the progression of the disease.
Changes in DNA methylation patterns can also be used to develop new treatments for diseases. For example, drugs targeting DNA methylation enzymes can reverse hypermethylation and restore regular gene expression, which may lead to the reversal of certain conditions.
Overall, changes in DNA methylation give us valuable insight into the underlying mechanisms of diseases and can significantly improve our understanding of and ability to treat a wide range of health conditions.
But it’s important to note that epigenetics is not just about the effects of lifestyle. Epigenetics refers to heritable changes in gene expression that do not involve changes to the underlying DNA sequence. While lifestyles and environmental factors, such as diet, exercise, and stress, can and often do play a role in shaping the epigenome and influencing gene expression, they are not the only factors contributing to epigenetic changes.
Pollutants in our air may influence epigenetic modifications. High levels of air pollution can have similar effects to that of occasional smoking. Pollutants in our water and food also have the potential to evoke epigenetic changes. For example, there are a number of recent studies showing high levels of “forever chemicals”, which can influence cell differentiation or even cause cancer, in fish or other common food items. Critically, unlike that for adverse exposures such as work stress or smoking, we often do not appreciate that we have been exposed to these chemicals. As a result, our understanding of the effect of these potentially harmful factors lacks the “granular” details necessary to develop an exacting knowledge the impact of these substances on our epigenome.
In summary, epigenetics is a complex and multifaceted field that involves the interplay between genetic, environmental, and lifestyle factors and is not limited to the influence of lifestyle alone.
References:
-
- Whole Genome Bisulfite Sequencing (WGBS) Applications – News-Medical.net. https://www.news-medical.net/life-sciences/Applications-of-Whole-Genome-Bisulfite-Sequencing-(WGBS).aspx
- Impact of DNA methylation on 3D genome structure. https://www.nature.com/articles/s41467-021-23142-8
- Gao, Yuan, et al. “Time-Course Transcriptome Analysis Reveals Resistance Genes of Panax Ginseng Induced by Cylindrocarpon Destructans Infection Using RNA-Seq.” PLoS One, vol. 11, no. 2, Public Library of Science, Feb. 2016, p. e0149408.
- Strobl, Barbara, et al. “The CrowdWater Game: A Playful Way to Improve the Accuracy of Crowdsourced Water Level Class Data.” PLoS One, vol. 14, no. 9, Public Library of Science, Sept. 2019, p. e0222579.
- Wang, Wei, et al. “Soil Fungal Biodiversity and Pathogen Identification of Rotten Disease in Aconitum Carmichaelii (Fuzi) Roots.” PLoS One, vol. 13, no. 10, Public Library of Science, Oct. 2018, p. e0205891.